Researchers from our consortium member CAM-SU GRC, Soochow University, have developed a machine-learning algorithm to identify candidate genes linked to disruption of circadian behaviour, eating behaviour and other cyclical biological processes. Using the algorithm, the researchers screened 750 different mouse mutant lines, finding five candidate genes linked to circadian misalignment.
What Are Circadian Rhythms?
Circadian rhythms, our daily biological processes and cycles like sleeping, eating and hormones, play a key role in our physical and psychological health. In a healthy individual, these cycles are regulated internally and externally in a few different ways. Externally, natural factors called ‘zeitgebers’, such as light, activity levels and melatonin, act as cues for our circadian rhythms so that they gradually synchronise to our environment. Internally, we have our suprachiasmatic nucleus (SCN) located in the brain which generates 24-hour rhythms. The SCN acts as a master clock, controlling vital rhythms such as our sleep/wake cycle, mood and temperature. Our organs and larger systems have their own circadian patterns and our cells have their own genetic and molecular clocks. These peripheral rhythms are regulated by the SCN via hormones, our nervous system and proteins in sync with day and night changes.
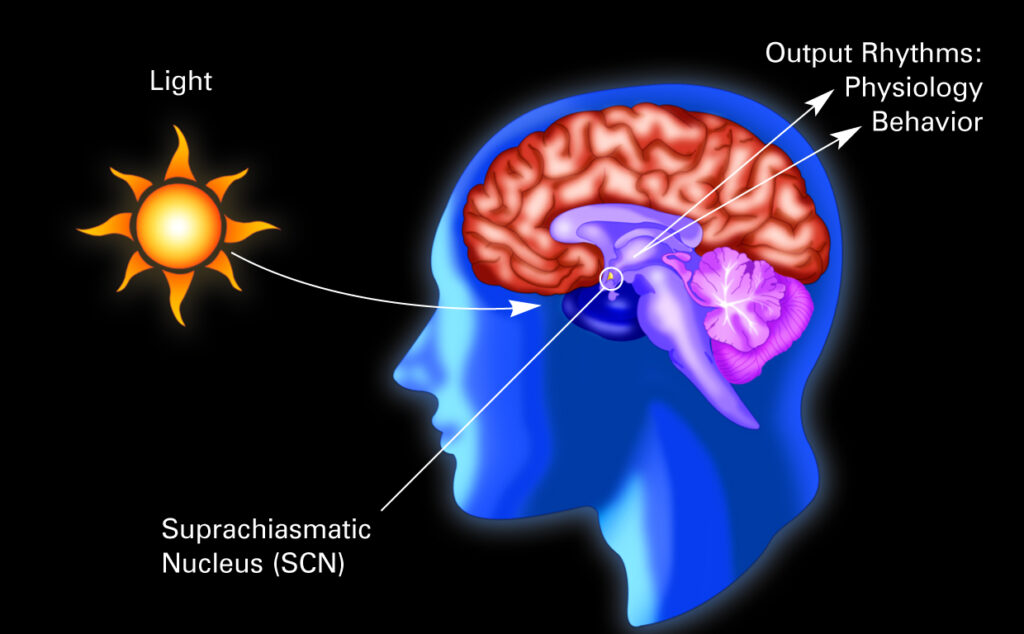
Credit: NIGMS
Circadian ‘misalignment’ is where our healthy 24-hour circadian rhythms become disrupted and unbalanced. Sometimes lifestyle can cause this, such as jet-lag and shift work, but this can also be caused by genetics. Misalignment between our internal clock and external factors can promote conditions like insomnia and other sleep disorders, as well as various psychiatric disorders like depression, seasonal affective disorder and bipolar disorder. Circadian misalignment is therefore crucial for our understanding of complex health issues.
Finding the Cause of Misalignment
A great way of finding new genes that can cause disorders and disease is by using bioinformatics to screen large databases, for example, creating new algorithms to find abnormalities in data. To create an algorithm which could find genes linked to circadian misalignment, the researchers first had to found the best measurable factors (parameters) to analyse in mice for circadian misalignment. They collected and used data from five IMPC partners: the Riken BioResource Center, the Centre for Phenogenomics, the Institut Clinique de la Souris, the Wellcome Trust Sanger Institute and Helmholtz Zentrum Munich. These centres collect ‘indirect calorimetry’ (IC) data – measures of metabolic rate (oxygen use, carbon dioxide production), food intake and locomotor activity – to provide insight into a mouse’s energy use, such as metabolism, eating, sleeping and physical activity. Using this data, they compared 750 mutant mouse types (where a single gene had been ‘switched off’, causing the gene to either partially or fully stop functioning) against 2,000 ‘wild-type’ mice – healthy mice with no mutations and a known genetic background.
The researchers analysed the IC data from the wild-type mice and found that both activity and food intake data had two clear peaks, which they labelled a strong early evening (E) peak and a weak early morning (M) peak. These were the reliable parameters they chose to compare the mutant mice against.
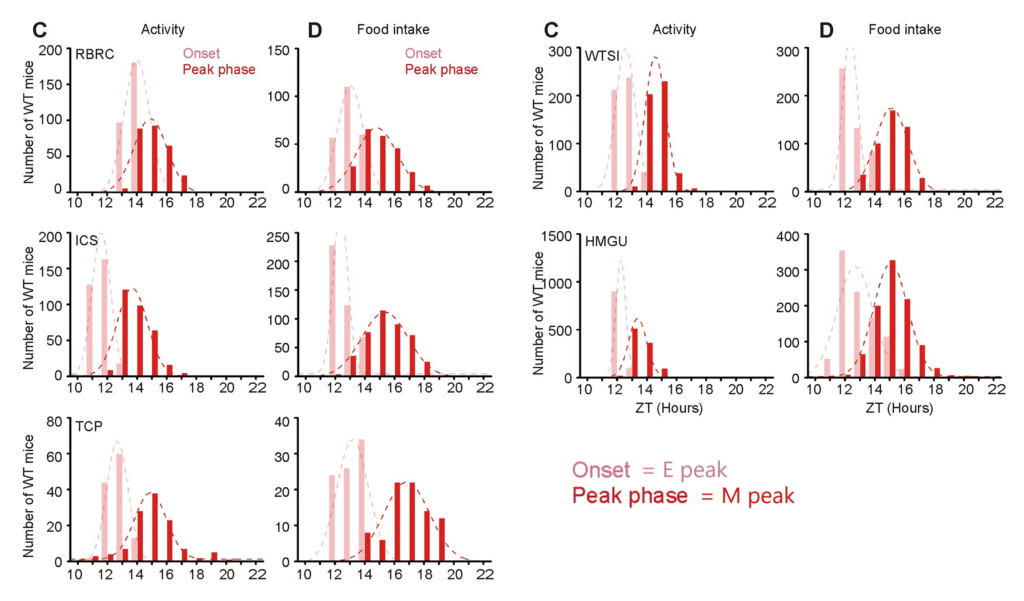
The algorithm learned how to analyse data and produce predictive models that could be used to find candidate genes linked to circadian misalignment. The researchers made synthetic datasets with labelled M and E peaks which they used to ‘train’ the machine-learning algorithm. They then tested it on real IC datasets and manually checked its accuracy to find a 95% agreement between the algorithm’s results and manually assessing the datasets. As a last accuracy and reliability step, they tested the algorithm on genes that are already known to cause circadian misalignment issues: hPER2S662G, Fbxl3KO, Nestin-cre;Zbtb20 and hPER1S714G mutants. These genes affect a mouse’s eating or activity patterns in different ways, for example, hPER2S662G mice have an advanced onset of activity, whilst hPER1S714G mice have an advanced onset of food intake. The machine learning algorithm successfully ‘flagged’ mice with mutated versions of these genes by accurately identifying the known abnormalities. This test showed the researchers that the algorithm could reliably evaluate mutant mouse phenotypes and recognise phenotypic abnormalities linked with circadian misalignment. Professor Ying Xu from CAM-SU, Soochow University, who was the lead in this international collaborative project, stresses:
“It’s important in mouse genetics to use an unbiased approach to better understand gene-environment interactions that could be related to human disease.”
Putting the Algorithm into Action
The initial screening of the 750 mutant mouse types flagged 88 genes that were possibly related to circadian misalignment. These 88 genes also had to meet three criteria: 50% of the mice in each mutation group had to have similar phenotypes, the difference between mutants and wild-type mice had to be statistically significant and the effect size had to be above 1.2, which further suggests statistical significance. Five mutant lines (Slc7a11tm1b/tm1b, Rhbdl1+/tm1.1, Spop+/tm1b, Oxtrtm1.1/tm1.1, Ctc1+/tm1b) matched all three criteria. The researchers chose to further test Slc7a11, which, when turned off, caused early activity onset and potentially impaired metabolism due to altered glucose tolerance. They generated Slc7a11 deficient mice and double-checked the phenotypes with a series of tests, such as jet-lag simulations and light sensitivity tests. These tests showed that the disruption of Slc7a11 affected the mice’s sensitivity to light/dark cycles. Further study of Slc7a11’s role in the SCN showed that, whilst it didn’t have a role in core clock function, Slc7a11 does play a role in SCN neuron synchronisation and messaging between cells – important cellular communication methods the body uses to regulate processes.
One of the corresponding authors, Dr. Jan Rozman, Czech Centre for Phenogenomics (Institute of Molecular Genetics, Czech Academy of Sciences), says:
“The paper resulted from an international collaboration of research institutions under the umbrella of IMPC. The findings highlight what can be achieved by joining forces in a global large-scale project. Future research is required to decipher the molecular and physiological functions of the identified candidate genes.”
This study has shown that large-scale analysis and characterisation of mouse mutants can highlight new candidate genes involved in circadian misalignment both easily and efficiently. The algorithm created, described and tested in this study can be used for future dataset analysis, opening new research possibilities into circadian misalignment for metabolism, sleep and behaviour. New research, such as the further investigation into the other four genes found in this initial screen, could improve our understanding of internal circadian clocks and provide insight into the growing number of conditions and disorders circadian misalignment is being connected to.
Source
Zhang T, Xie P, Dong Y, Liu Z, Zhou F, Pan D, et al. (2020) High-throughput discovery of genetic determinants of circadian misalignment. PLoS Genet 16(1): e1008577. https://doi.org/10.1371/journal.pgen.1008577

